Abstract
Purpose- Experimental models of hemophagocytic lymphohistiocytosis (HLH) have revealed a disorder characterized by cytotoxic T cell activation and driven by IFN-g (Jordan, Hildeman et al. Blood, 2004). Recently, a multiplex protein array of 135 proteins in human plasma demonstrated that IFN-g signaling in HLH is uniquely elevated compared to sepsis (Lin, Scull, et al. Blood advances, 2021). However, to date, broader proteomic methods have not been utilized to study familial HLH (FHL). Therefore, we aimed to study the biological pathways of familial HLH with an unbiased approach and broadly characterize the inflammation seen in these patients.
Methods- We conducted both proteomic and gene expression studies examining the serum and peripheral blood mononuclear cells (PBMCs), respectively, of patients with HLH. We utilized the SomaScan platform (SomaLogic) to assay 7,596 proteins in serum and bulk RNAseq to assay gene expression in PBMC. The proteomic study included seven FHL patients treated at Schneider Children's Medical Center of Israel and seven age-matched controls (ages five weeks- 58 months). Biallelic genetic lesions were identified in five patients, while two had no known genetic defects. RNAseq studies included six familial HLH patients (four with biallelic genetic lesions) and five healthy pediatric controls from Cincinnati Children's Hospital Medical Center (CCHMC) and were conducted at CCHMC's RNAseq core. For unsupervised clustering of patient results, primary component analysis, t-SNE, and heatmap analyses were performed. In addition, differentially expressed proteins and genes were compared to cross-validate and define common proteins/genes between proteomics and RNAseq. Finally, gene cell enrichment analysis (GSEA) with hallmark pathways was performed on the intersecting genes/proteins to identify the significant pathways identified with each modality.
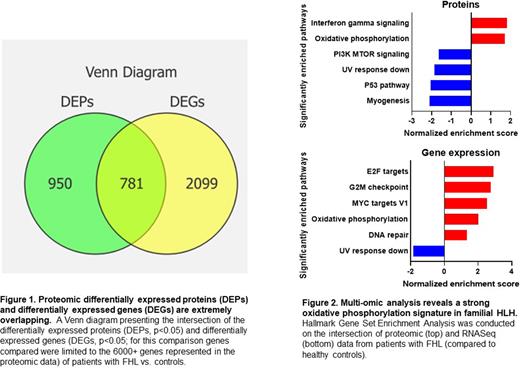
Results- FHL samples readily clustered separately from aged-matched pediatric controls (CTL) in our gene expression and proteomic assays. Overall, 1,731 proteins were differential (p<0.05) and 10,036 genes were differential (p<0.05) when comparing FHL and CTL groups. Of the differential genes identified by RNASeq, 2,880 were represented in the proteomic assay. Of these, 781 genes/proteins were significantly differential by both modalities (Figure 1). GSEA pathway analysis of the intersecting genes revealed that while IFN-g signaling was uniquely enriched in the proteomics (Figure 2, normalized enrichment score 1.83, p=0.01, q=0.23) oxidative phosphorylation was highly enriched in both platforms (proteomics normalized enrichment score 1.72, p=0.02, q=0.22, Figure 2B; RNAseq normalized enrichment score 2.0, p=0.005, q=0.009, Figure 2). Finally, we created a multi-omic validated set of differential proteins through this intersection analysis.
Conclusion- We report the first deep multi-omic analysis of FHL compared to healthy pediatric controls that validate the importance of IFN-g in the disease pathophysiology and highlight an unexpected signature of oxidative phosphorylation as an important differential pathway. As both activated T cells and M1 macrophages are thought to rely primarily on glycolysis, the source of this oxidative phosphorylation is unclear. These findings support prior findings from experimental models and previous limited cytokine studies while also delineating new unbiased discoveries in this unique disorder. More extensive proteomic characterization may identify novel pathways for therapeutic targeting in HLH.
Disclosures
Jordan:Sobi, Inc.: Consultancy, Research Funding. Zoref-Lorenz:Sobi: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal